
by Hélène Rivière-Rolland, Loïc Taloc, Danielle Ziébelin, François Rechenmann and Alain Viari
A knowledge base to represent metabolism data has been developed by the Helix research team at INRIA Rhône-Alpes. This base provides access to information on chemical compounds, biochemical reactions, enzymes, genes and metabolic pathways from fully sequenced micro-organisms. The model has been implemented by using an object/association technology developed at INRIA. Beside its use as a general repository, the base may have applications in metabolic simulations and pathway reconstruction in newly sequenced genomes.
The cellular metabolism can be defined as the panel of all biochemical reactions occurring in the cell. It consists of molecular synthesis (anabolism) and degradation (catabolism) necessary for cell growth and division. These reactions drive the energetic processes, the synthesis of structural and catalytic components of the cell and the elimination of cellular wastes.
A fairly large amount of metabolic data is readily available, either in the literature or in public data banks (eg the KEGG project: http://star.scl.genome.ad.jp/kegg) and this information will probably grow in the near future due to the development of new ‘large scale’ experimental technologies like DNA-arrays. Therefore, there is a need to organise this data in a rational and formalised way, ie to model our knowledge of metabolic data. The first goal is of course the storage and recovery of pertinent information. The complexity of this kind of data and in particular the fact that some information is held in the relationship between the biological entities rather than in the entities themselves, makes their selection and recovery difficult. Moreover, our knowledge in this area is often incomplete (elements are missing or pathways may be totally unknown in a newly sequenced organism). A challenge is therefore to cope with this partial information and to develop databases that could provide some inference mechanisms to assist the discovery process. Finally, another challenge is to link these data to other relevant genomic and biochemical information like protein structure, regulation of gene expression, whole genome organisation (eg syntheny) and evolution.
Following the pioneering work of P. Karp and M. Riley with the Eco-Cyc system (http://ecocyc.PangeaSystems.com/ecocyc) we attempted to develop a knowledge base of metabolic data. We wanted to experiment a different representation model in which associations are explicitly represented as entities. To this purpose, we used the AROM system developed at INRIA (http://www.inrialpes.fr/ romans/ pub/arom). The main originality of AROM is the presence of two complementary entities of representation: classes and associations. As in any object-oriented system, a class represents a set of objects described by slots; but, in AROM, such a slot cannot refer to another object. This connection is done by means of associations which therefore denotes a set of tuples (not necessarily only two) of objects (associations are therefore n-ary). As objects, tuples have their own slots and as classes, associations can be organised in hierarchies therefore allowing for usual inheritance and specialisation mechanisms. The explicit representation of n-ary associations turned out to be very useful for representing biological data. For instance, it makes the representation of alternative substrates of a metabolic reaction a much easier task.

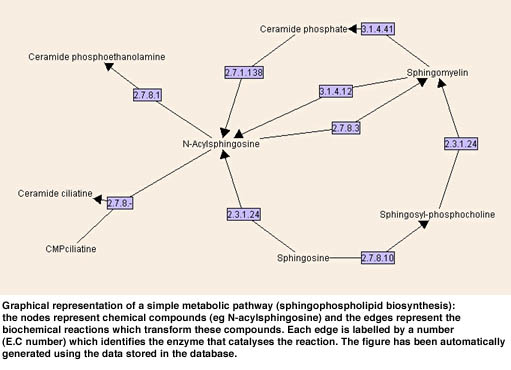
After implementing the data model in AROM, we extracted the metabolic data from public sources (mostly KEGG) by using parsers and Unix shell scripts. Coherence of sequence data between data banks has been checked by using home-made sequences alignment programs and/or Blast. At the present time we are developing several graphical interfaces to this base. One will be devoted to querying the knowledge base. Another interface will be devoted to the automatic graphical representation of pathways which are complex non-planar directed graphs (see Figure). At the present time all the system (AROM and the inter faces) is implemented in JAVA and we plan to put it into play through a web applet-server in a near future.
Links:
Action Helix: http://www.inrialpes.fr/helix.html
Please contact:
Alain Viari - INRIA
Tel: +33 4 76 61 54 74
E-mail: alain.viari@inrialpes.fr