Mining Distributed and Heterogeneous Clinical Data Sources
by George Potamias
The HealthObs (Health Observatory) integrated environment offers seamless integration and intelligent processing of distributed and heterogeneous clinical information. This is achieved through the use of XML and data mining. Its aim is to assist healthcare professionals in coming to grips with the vast amounts of information and to enhance their decision-making capabilities.
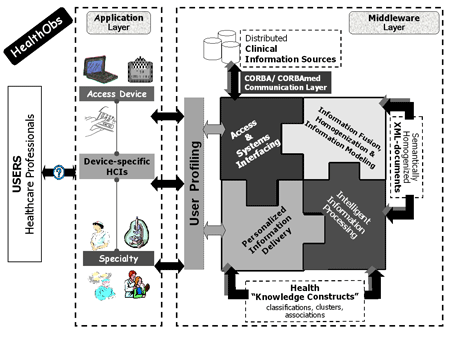
HealthObs is an info-mediation and brokerage environment with 'knowledge discovery' functionality. It is composed of two synergistic layers (see Figure 1).
The Middleware Layer is a set of software components that enables (i) access to the distributed information and data sources regardless of platform, location and type of information system; (ii) fusion and semantic homogenization of information and data items, enabled by the uniform information modelling of the underlying information and data items and supported by advanced ontology and RDF/XML technologies; and (iii) intelligent information and data processing based on advanced data mining operations.
|
The Application Layer is based on the use and integration of the middleware components, for specific medical domains.
Central to the architecture is a single data-enriched XML file that contains information and data from remote (and potentially heterogeneous) clinical information systems. To this end, the Integrated Electronic Health Care Record (IEHCR) services of HYGEIAnet (the Regional Health Information Network (RHIN) of Crete; http://www.hygeianet.gr) are utilized in order to query the federated clinical information sources and recall the relevant query-specific data items. For each query, and with the aid of custom-made filtering and formatting operations, a query-specific XML file is created. HealthObs initiates and bases its knowledge discovery (ie association rules mining) operations on such data-enriched XML files. In this respect, HealthObs falls into the category of XML-content mining tools.
Semantic Homogenization and Domain Adaptation
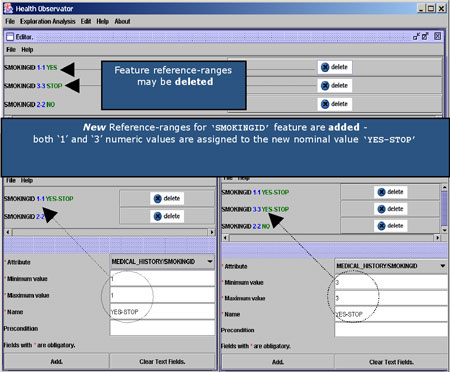
A domain-specific ontology is required in order to hide the heterogeneous nature of data. A special service known as the Common Clinical Term Reference service (CCTR) has been developed for the storage and retrieval of common and universally accepted names and codes of medical terms. This service uses terms and relations from the ICD9 (International Coding of Diseases) and ICPC (International Classification of Primary Care) standard coding schemes. Moreover, when dealing with clinical laboratory findings it is crucial to refer to qualitative, rather than numeric measurements. CCTR offers a means of assigning qualitative values to the different measurements. In HealthObs this is made operational via a special Domain Editor tool (see Figure 2).
|
Mining Interesting Clinical Associations
To enable the adaptation of Association Rules Mining (ARM) operations we have instituted two key conventions. In the first of these, each transaction corresponds to a specific patient encounter (ie identifiable visits of patients to a healthcare unit within the federation). Each encounter/visit is uniquely identified by reference to three attributes recorded in the respective clinical information systems; namely patientid, information-system-id, and encounter-id (or visit-id). Patient data are recalled anonymously and retrieved in a secure manner by security and role-based authorization services offered by the RHIN information infrastructure. In the second convention, each item is represented by the following triplet: '<SyntheticFeature, AtomicFeature, AtomicFeatureValue>', the entries of which correspond to instance elements present in the XML file to be processed. For example, a SyntheticFeature may correspond to the 'biochemical test' element; AtomicFeature to an element that stands for a specific biochemical test, eg 'glucose'; and FeatureValue to the value of the atomic feature, eg '68.0' for glucose.
With the aid of the ARM component, HealthObs is able to form significant and useful associations of the type: X1, X2, ... Xn ' Y1,Y2, ..., Ym, where each pair <Xi, Yi> corresponds to specific clinical features and findings. For example, "IF GLUCOSE = high and UREA = high THENISCHEMIC-HEART disease [Support: 1%, Confidence: 60%]."
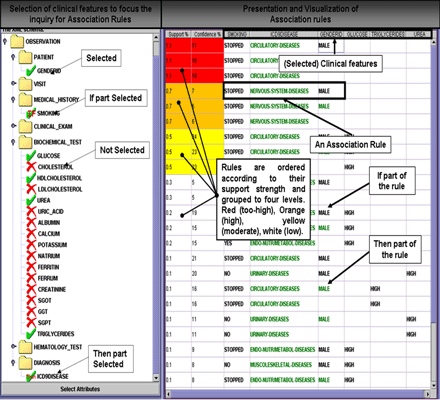
In HealthObs, both the selection of clinical features to focus the knowledge discovery operations, and the visualization of discovered association rules are achieved via a specially designed graphical user-interface (GUI). An indicative screen-shot of this interface is shown in Figure 3.
|
HealthObs system has been successfully used for various knowledge discovery tasks in the context of the HYGEIAnet RHIN. Furthermore, work is in progress to extend the system's information and data model towards the incorporation and management of genomic information (eg patients' samples, gene-expression profiles, gene markers etc). The work is being undertaken in the context of the PrognoChip project funded by the Greek Secretariat for Research & Technology, the aim of which is the discovery of reliable prognostic molecular (ie gene) markers for breast cancer and their linkage and validation with identifiable clinico-histological patient profiles.
Please contact:
George Potamias, ICS-FORTH, Greece
E-mail: potamias![]() ics.forth.gr
ics.forth.gr

 This issue in
This issue in